Source Codes are available upon requests
3D Massive Blobs Detection(Unsupervised&Supervised) Machine Learning in Medical Imaging
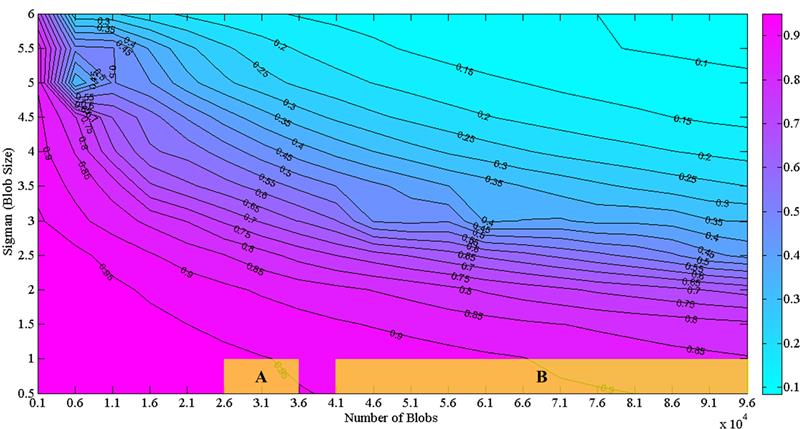
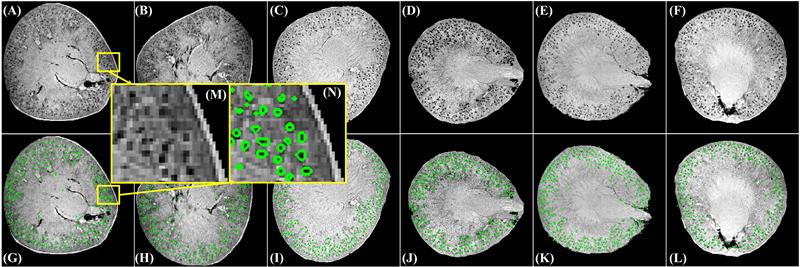
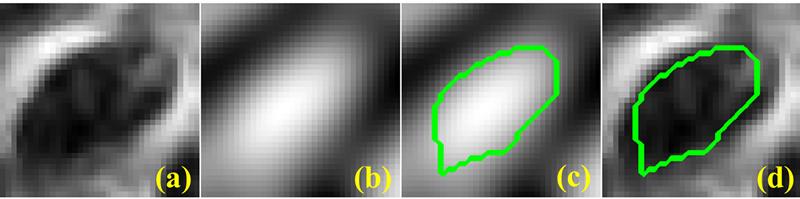
Magnetic resonance imaing (MRI) is a powerful tool for noninvasive structural and molecular investigation of tissue. Targeted molecular imaging contrast agents are making it possible to localize molecules and cells throughout the body, potentially offering a three-dimensional view of disease progression and response to therapy. Superparamagnetic contrast agents, often formed of nanoparticles, have long been proposed as a tool to deliver contrast to a specific site or to track jndivdual or multiple cells in the body. However, detecting these agents against a variable image background is difficult, and most of the proposed techniques to segment targeted structures have been implemented only in two dimensions. Here we propose an efficient Hessian-based Difference of Gaussians (HDoG) detector to segment glomeruli on 3D MR images enhanced by superparamagnetic iron oxide contrast agents. Specifically, we tested the technique to detect labeled renal glomeruli in 3D in both rat and human kidneys after injection of the iron oxide contrast agent cationized ferritin. HDoG is based on the scale-space theory, where the Gaussian scale-space representation of an image is defined as a convolution of the raw images over a Gaussian kernel given a scale parameter, t. As with other similar detectors, we first smoothed the image with the standard difference of Gaussians (DoG) algorithm to identify all potential glomeruli. Then, the Hessian analysis pre-segmented and outlined the candidate glomeruli. Next, we determined regional features associated with each candidate and performed post-pruning using an unsupervised clustering algorithm, eliminating any false glomeruli and producing the final segmentation. To test the HDoG algorithm, we used specific metrics to compare the performance of HDoG with that of three other detectors (LoG, gLoG and HLoG) when applied to 2D images. HDoG outperformed the three detectors in identifying structures of interest on the 2D images, and it was more computationally efficient. Finally, we examined how well the HDoG segmented glomeruli on 3D MR images of kidneys; we imaged six rat and three human kidneys. Overall, HDoG performed as well as existing histological techniques for the same measurements.
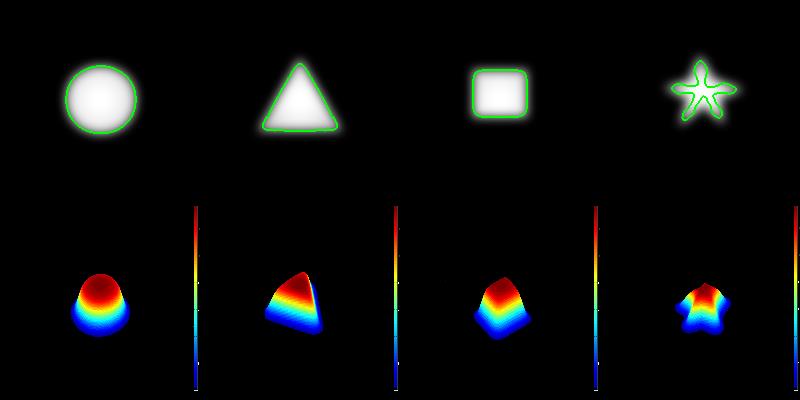
Publication
Min Zhang, Teresa Wu, Scott Beeman, Luise Cullen-McEwen, John Bertram, Jennifer Charlton, Edwin Baldelomar, Kevin Bennett: Efficient Small Blob Detection based on Local Convexity, Intensity and Shape Information. IEEE Transactions on Medical Imaging vol. 35, no. 4, pp. 1127-1137, April 2016. doi: 10.1109/TMI.2015.2509463
Min Zhang, Teresa. Wu, Kevin.M. Bennett: A novel Hessian based algorithm for rat kidney glomerulus detection in 3D MRI. Proceedings of SPIE Medical Imaging; 03/2015; DOI: 10.1117/12.2081484
Unsupervised Microstructure (Cell) Segmentation Machine Learning in Medical Imaging
Recent advances in medical imaging technology have greatly enhanced imaging based diagnosis which requires computational effective and accurate algorithms to process the images (e.g., measure the objects) for quantitative assessment. In this research, we are interested in one type of imaging object: small blobs. Examples of small blob objects are cells in histopathology images, glomeruli in MR images, etc. This problem is particularly challenging because the small blobs often have inhomogeneous intensity distribution and an indistinct boundary against the background. Yet, in general, these blobs have similar sizes. Motived by this finding, we propose a novel detector termed Hessian-based Laplacian of Gaussian (HLoG) using the scale space theory as the foundation. Like most imaging detectors, an image is first smoothed via LoG. Hessian analysis is then launched to identify the single optimal scale based on which a pre-segmentation is conducted. The advantage of the Hessian process is it is capable of delineating the blobs. As a result, regional features can be retrieved. These features enable the unsupervised clustering algorithm for post-pruning which shall be more robust and sensitive than the traditional threshold-based post-pruning commonly used in most imaging detectors. To test the performance of the proposed HLoG, two sets of 2D grey medical images are studied. HLoG is compared against three state-of-the-art detectors: gLoG, Radial-Symmetry and LoG using precision, recall and F-score metrics. We observe that HLoG statistically outperforms the compared detectors.
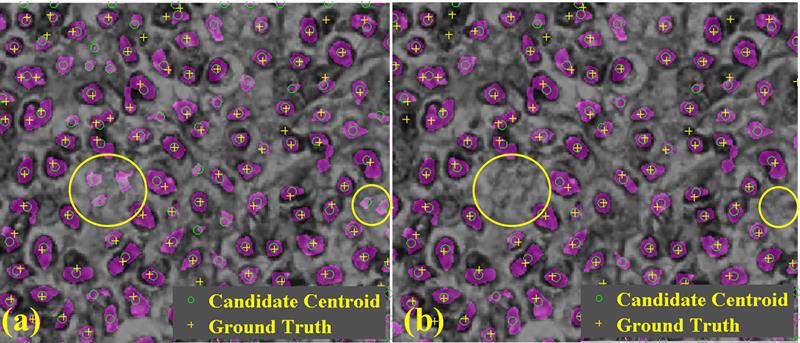
Publication
Min Zhang, Teresa Wu, Kevin M. Bennett: Small Blob Identification in Medical Images Using Regional Features From Optimum Scale. IEEE transactions on Biomedical Engineering 09/2014; 62(4). DOI:10.1109/TBME.2014.2360154
DICOM Index Tracker Healthcare Informatics
DICOM Index Tracker (DIT) is an integrated platform to harvest rich information available from Digital Imaging and Communications in Medicine (DICOM) to improve quality assurance in radiology practices. It is designed to capture and maintain longitudinal patient-specific exam indices of interests for all diagnostic and procedural uses of imaging modalities. Thus, it effectively serves as a quality assurance and patient safety monitoring tool. The foundation of DIT is an intelligent database system which stores the information accepted and parsed via a DICOM receiver and parser. The database system enables the basic dosimetry analysis. The success of DIT implementation at Mayo Clinic Arizona calls for the DIT deployment at the enterprise level which requires significant improvements. First, for geographically distributed multi-site implementation, the first bottleneck is the communication (network) delay; the second is the scalability of the DICOM parser to handle the large volume of exams from different sites. To address this issue, DICOM receiver and parser are separated and decentralized by site. To facilitate the enterprise wide Quality Assurance (QA), a notable challenge is the great diversities of manufacturers, modalities and software versions, as the solution DIT Enterprise provides the standardization tool for device naming, protocol naming, physician naming across sites. Thirdly, advanced analytic engines are implemented online which support the proactive QA in DIT Enterprise.
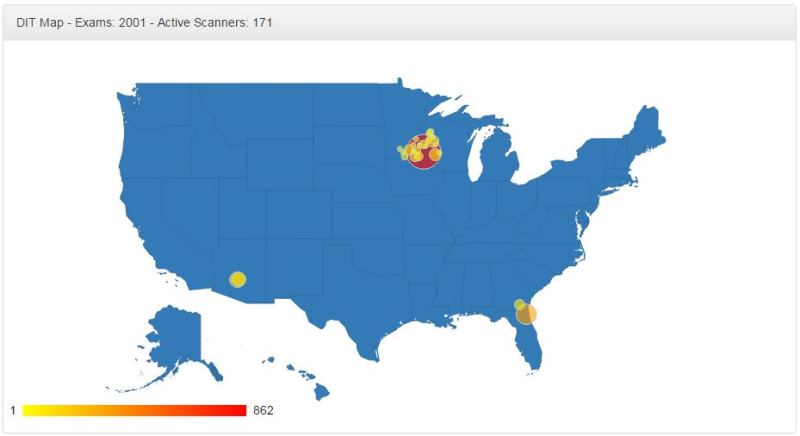
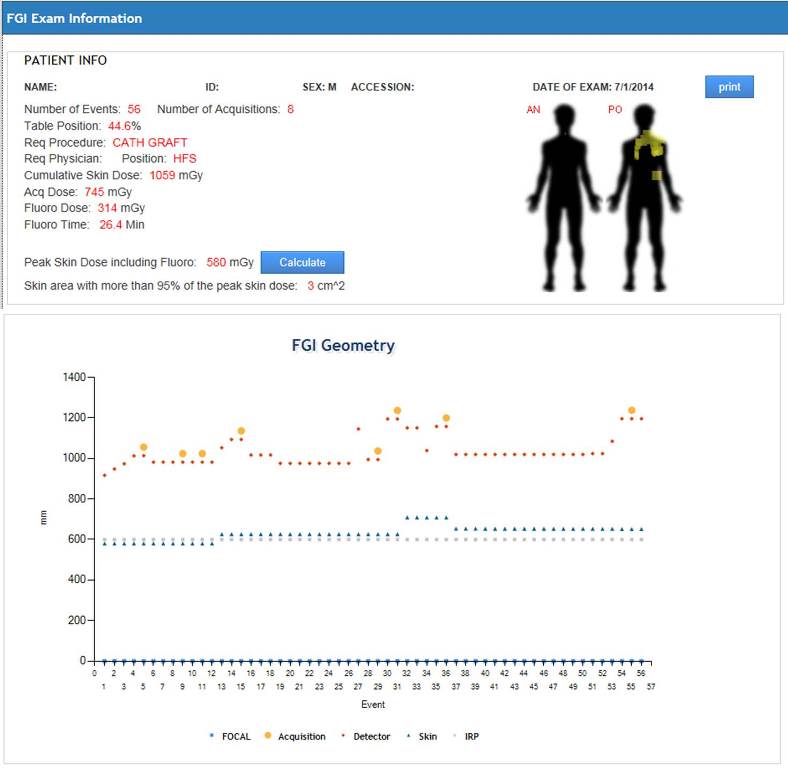
Publication
Min Zhang, William Pavlicek, Anshuman Panda, Steve G. Langer, Richard Morin, Kenneth A. Fetterly, Robert Paden, James Hanson, Lin-Wei Wu, Teresa Wu: DICOM index tracker enterprise: advanced system for enterprise-wide quality assurance and patient safety monitoring. Proceedings of SPIE Medical Imaging 03/2015; DOI:10.1117/12.2082120
Min Zhang, Clinton Wellnitz, Can Cui, William Pavlicek, Teresa Wu: Automated Detection of Z-Axis Coverage with Abdomen-Pelvis Computed Tomography Examinations. Journal of Digital Imaging; June 2015, Volume 28, Issue 3, pp 362-367. DOI:10.1007/s10278-014-9743-7
Mouse Brain Segmentation Machine Learning in Medical Imaging
We introduce an automated method, called prior feature Support Vector Machine- Markov Random Field (pSVMRF), to segment three-dimensional mouse brain Magnetic Resonance Microscopy (MRM) images. Our earlier work, extended MRF (eMRF) integrated Support Vector Machine (SVM) and Markov Random Field (MRF) approaches, leading to improved segmentation accuracy; however, the computation of eMRF is very expensive, which may limit its performance on segmentation and robustness. In this study pSVMRF reduces training and testing time for SVM, while boosting segmentation performance. Unlike the eMRF approach, where MR intensity information and location priors are linearly combined, pSVMRF combines this information in a nonlinear fashion, and enhances the discriminative ability of the algorithm. We validate the proposed method using MR imaging of unstained and actively stained mouse brain specimens, and compare segmentation accuracy with two existing methods: eMRF and MRF. C57BL/6 mice are used for training and testing, using cross validation. For formalin fixed C57BL/6 specimens, pSVMRF outperforms both eMRF and MRF. The segmentation accuracy for C57BL/6 brains, stained or not, was similar for larger structures like hippocampus and caudate putamen, (~87%), but increased substantially for smaller regions like susbtantia nigra (from 78.36% to 91.55%), and anterior commissure (from ~50% to ~80%). To test segmentation robustness against increased anatomical variability we add two strains, BXD29 and a transgenic mouse model of Alzheimers Disease. Segmentation accuracy for new strains is 80% for hippocampus, and caudate putamen, indicating that pSVMRF is a promising approach for phenotyping mouse models of human brain disorders.
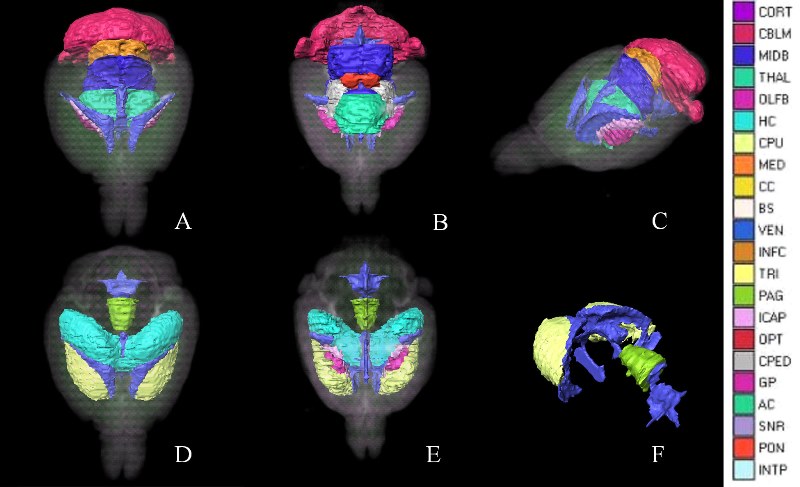
Publication
Teresa Wu, Min Hyeok Bae, Min Zhang, Rong Pan, Alexandra Badea: A prior feature SVM-MRF based method for mouse brain segmentation. NeuroImage 02/2012; 59(3):2298-306. DOI:10.1016/j.Neuroimage.2011.09.053